We use an integrated structural biology and biophysical approach combined with functional studies to understand the inhibition mechanisms of novel protein–RNA targets involved in human diseases and translate it to develop novel therapeutics.
Background and Current Research
We study the molecular mechanisms of protein-RNA complexes and their applications for the development of novel inhibitors. We are interested in:
– AminoAcyl-tRNA Synthetases (AARS);
– 3’end mRNA Processing Factors and Polyadenylation;
– 3’end pre-tRNA Processing.
AminoAcyl-tRNA Synthetases (AARS), a family of proteins responsible for attaching the correct amino acid to the cognate tRNA, a process called aminoacylation; and preventing mischarging of tRNAs to non-correct amino acids (editing). AARSs are interesting therapeutic targets and housekeeping proteins that: i) play a fundamental role for protein translation, a critical process during fast growing conditions of microbes; and ii) present differences between microbes and humans that can be used for drug discovery.
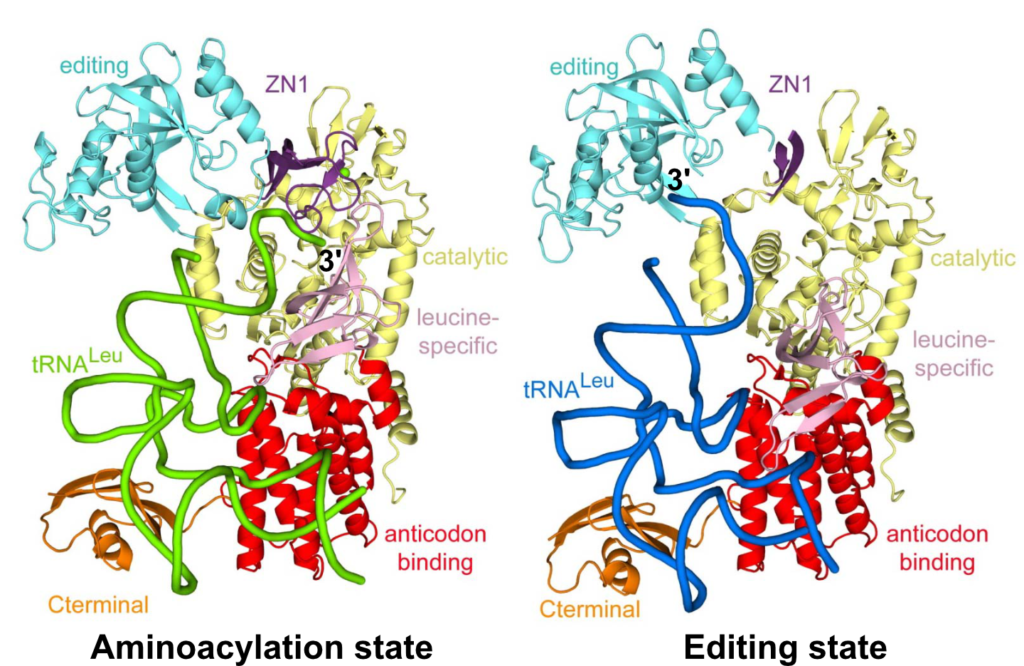
We focus on Leucyl-tRNA synthetase (LeuRS), which charges leucine to the cognate tRNA and prevents mischarging to related amino acids. In 2012 we published structures of LeuRS in different functional states: 1) the aminoacylation state, where the 3’-tRNA is in the synthetic site ready to accept leu-AMP; 2) the post-transfer editing state, where the mischarged 3’-tRNA is bound into the editing site for proof-reading. Our structures provided unprecedented insight into the structural dynamics and the translocation of the 3’-tRNA between the synthetic and the editing sites, separated > 35 Å (Palencia et al. 2012, Chopra et al. 2013) (Fig1).
Please see video here.
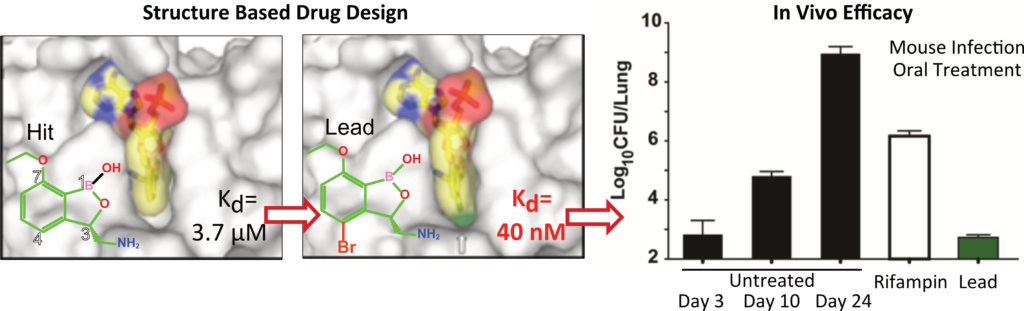
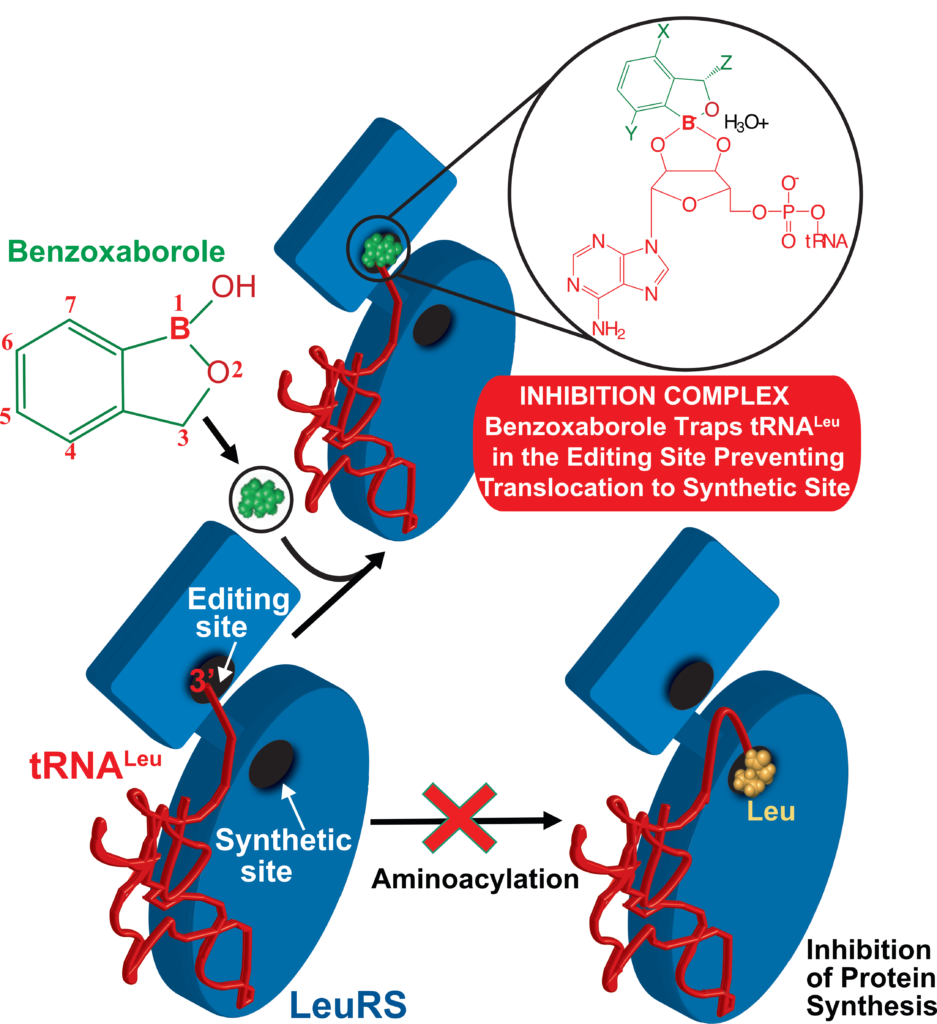
Using this mechanistic insight, in close collaboration with multidisciplinary teams in academia and industry, we contributed to the development of novel antimicrobials targeting LeuRS. Our structures of LeuRS–tRNA bound to benzoxaboroles unveiled the inhibition mechanism of novel antibacterials, whereby the tRNA gets trapped at the editing site and effectively inhibits protein synthesis (Fig2). Using a structure-based drug design approach we brought the potency of initial hits from micro- to nano-molar and improve selectivity, allowing their progression into clinical trials. Exploiting this mechanism of action, we also discovered novel antiinfectives against apicomplexan parasites.
An exciting area of our current research is the targeting of other protein-RNA complexes such as cleavage and polyadenylation specificity factors (CPSF). In a highly interdisciplinary international collaboration we discovered a novel benzoxaborole inhibitor of eukaryotic parasites that targets CPSF3 (Palencia et al. 2017, Sonoiki et al. 2018). In eukaryotes, most of the 3’ pre-mRNA processing occurs via the concerted action of a multiprotein complex that cleaves the 3’-end of unmatured mRNAs before the addition of the poly-A tail and their export to the cytoplasm for protein translation. CPSF3 has a key function within this complex as it is the endonuclease responsible of cleaving 3’ pre-mRNAs.
Future directions and goals
We will provide new structural insights into the inhibition of LeuRS, and in collaboration with other academic groups and industry, we will improve the potency and selectivity of inhibitors with biomedical applications. A main focus will be the improvement of current antibacterial inhibitors in order to minimize the frequency of drug resistance.
We will expand this approach to other recently discovered drug targets, notably CPSF3, to understand its inhibition mechanism and to guide medicinal chemistry on the design of inhibitors with improved properties. Finally, there is emerging evidence showing that the homologous proteins of these pathogen’s drug targets in humans are deregulated and can lead to diseases. We will complement our structural/biophysical work with the expertise on bioinformatics, cell biology and relevant animal models available at our institute or abroad, to explore whether inhibition of these proteins can have therapeutic applications in human diseases, notably in cancer.
Selected Publications
– Targeting a microbiota Wolbachian aminoacyl-tRNA synthetase to block its pathogenic host. Science Adv. 10(28) 10.1126/sciadv.ado1453 (2024).
– Hoffmann, G., Le Gorrec, M., Mestdach, E., Cusack, S., Salmon, L., *Jensen, MR., *Palencia, A. Adenosine-Dependent Activation Mechanism of Prodrugs Targeting an Aminoacyl-tRNA Synthetase. Journal of the American Chemical Society. 10.1021/jacs.2c04808 (2023).
– Mariño Perez, L., Ielasi, F.S., Bessa, L.M., Maurin, D., Kragelj, J., Blackledge, M., Salvi, N., Bouvignies, G., *Palencia, A., *Jensen, M.R. Visualizing protein breathing motions associated with aromatic ring flipping. Nature. 602, 695-700 (2022).
– Ielasi, FS., Ternifi, S., Fontaine, E., Iuso, D., Couté, Y. & *A. Palencia. Human Histone pre-mRNA Assembles Histone- or Canonical-mRNA Processing Complexes by Overlapping 3’-End Sequence Elements. Nucleic Acids Research. Nov 30;gkac878 (2022).
– Lukarska, M. & *Palencia, A. Aminoacyl-tRNA synthetases as drug targets. The Enzymes, Elsevier. 48:321-350. doi: 10.1016 (2020).
– Swale, C., Bougdour, A., Gnahoui-David, A., Tottey, J., Georgeault, S., *Laurent, F., *†Palencia, A., *†Hakimi, M.A. Metal-captured inhibition of pre-mRNA processing activity by CPSF3 controls Cryptosporidium infection. Science Trans. Medicine Nov 6;11(517). doi: 10.1126 (2019). †Lead Authors.
– *Palencia, A., *Bougdour, A., et al. Targeting Toxoplasma gondii CPSF3 as a new approach to control toxoplasmosis. EMBO Molecular Medicine 9: 385-394 (2017). Highlighted in: France 3 news.
– Chopra, S.†, Palencia, A.†, et al. Structural characterisation of antibiotic self-immunity tRNA synthetase in plant tumour biocontrol agent. Nature comm 7:12928 (2016).
– Palencia, A.†,Chopra, S.†, et al. Plant tumour biocontrol agent employs a tRNA-dependent mechanism to inhibit leucyl-tRNA synthetase. Nature comm
4, 1417 (2013).
– Palencia, A.†, Crepin, T.†, et al. Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nature Struct. & Mol. Biol. 19, 677-84 (2012).
Palencia’s Group, Institute for Advanced Biosciences (IAB), Inserm U1209,
CNRS UMR5309/ Univ. Grenoble-Alpes, Grenoble
Contact: andres.palencia [ at] univ-grenoble-alpes.fr